netbet sports betting app

New technique reveals many possible conformations that a protein may take.
Anne Trafton | MIT News Office
February 4, 2021
Cryo-electron microscopy (cryo-EM) allows scientists to produce high-resolution, three-dimensional images of tiny molecules such as proteins. This technique works best for imaging proteins that exist in only one conformation, but MIT researchers have now developed a machine-learning algorithm that helps them identify multiple possible structures that a protein can take.
Unlike AI techniques that aim to predict protein structure from sequence data alone, protein structure can also be experimentally determined using cryo-EM, which produces hundreds of thousands, or even millions, of two-dimensional images of protein samples frozen in a thin layer of ice. Computer algorithms then piece together these images, taken from different angles, into a three-dimensional representation of the protein in a process termed reconstruction.
In a Nature Methods paper, the MIT researchers report a new AI-based software for reconstructing multiple structures and motions of the imaged protein — a major goal in the protein science community. Instead of using the traditional representation of protein structure as electron-scattering intensities on a 3D lattice, which is impractical for modeling multiple structures, the researchers introduced a new neural network architecture that can efficiently generate the full ensemble of structures in a single model.
“With the broad representation power of neural networks, we can extract structural information from noisy images and visualize detailed movements of macromolecular machines,” says Ellen Zhong, an MIT graduate student and the lead author of the paper.
With their software, they discovered protein motions from imaging datasets where only a single static 3D structure was originally identified. They also visualized large-scale flexible motions of the spliceosome — a protein complex that coordinates the splicing of the protein coding sequences of transcribed RNA.
“Our idea was to try to use machine-learning techniques to better capture the underlying structural heterogeneity, and to allow us to inspect the variety of structural states that are present in a sample,” says Joseph Davis, the Whitehead Career Development Assistant Professor in MIT’s Department of Biology.
Davis and Bonnie Berger, the Simons Professor of Mathematics at MIT and head of the Computation and Biology group at the Computer Science and Artificial Intelligence Laboratory, are the senior authors of the study, which appears today in Nature Methods. MIT postdoc Tristan Bepler is also an author of the paper.
netbet sports betting
The researchers demonstrated the utility of their new approach by analyzing structures that form during the process of assembling ribosomes — the cell organelles responsible for reading messenger RNA and translating it into proteins. Davis began studying the structure of ribosomes while a postdoc at the Scripps Research Institute. Ribosomes have two major subunits, each of which contains many individual proteins that are assembled in a multistep process.
To study the steps of ribosome assembly in detail, Davis stalled the process at different points and then took electron microscope images of the resulting structures. At some points, blocking assembly resulted in accumulation of just a single structure, suggesting that there is only one way for that step to occur. However, blocking other points resulted in many different structures, suggesting that the assembly could occur in a variety of ways.
Because some of these experiments generated so many different protein structures, traditional cryo-EM reconstruction tools did not work well to determine what those structures were.
“In general, it’s an extremely challenging problem to try to figure out how many states you have when you have a mixture of particles,” Davis says.
After starting his lab at MIT in 2017, he teamed up with Berger to use machine learning to develop a model that can use the two-dimensional images produced by cryo-EM to generate all of the three-dimensional structures found in the original sample.
In the new Nature Methods study, the researchers demonstrated the power of the technique by using it to identify a new ribosomal state that hadn’t been seen before. Previous studies had suggested that as a ribosome is assembled, large structural elements, which are akin to the foundation for a building, form first. Only after this foundation is formed are the “active sites” of the ribosome, which read messenger RNA and synthesize proteins, added to the structure.
In the new study, however, the researchers found that in a very small subset of ribosomes, about 1 percent, a structure that is normally added at the end actually appears before assembly of the foundation. To account for that, Davis hypothesizes that it might be too energetically expensive for cells to ensure that every single ribosome is assembled in the correct order.
“The cells are likely evolved to find a balance between what they can tolerate, which is maybe a small percentage of these types of potentially deleterious structures, and what it would cost to completely remove them from the assembly pathway,” he says.
Viral proteins
The researchers are now using this technique to study the coronavirus spike protein, which is the viral protein that binds to receptors on human cells and allows them to enter cells. The receptor binding domain (RBD) of the spike protein has three subunits, each of which can point either up or down.
“For me, watching the pandemic unfold over the past year has emphasized how important front-line antiviral drugs will be in battling similar viruses, which are likely to emerge in the future. As we start to think about how one might develop small molecule compounds to force all of the RBDs into the ‘down’ state so that they can’t interact with human cells, understanding exactly what the ‘up’ state looks like and how much conformational flexibility there is will be informative for drug design. We hope our new technique can reveal these sorts of structural details,” Davis says.
The research was funded by the National Science Foundation Graduate Research Fellowship Program, the National Institutes of Health, and the MIT Jameel Clinic for Machine Learning and Health. This work was supported by MIT Satori computation cluster hosted at the MGHPCC.

Thirteen postdocs and research scientists honored for contributions to the Institute with awards formerly known as Infinite Kilometer.
School of Science
February 3, 2021
This year, the MIT School of Science has recognized 13 postdocs and research scientists who are the recipients of the 2021 Infinite Expansion Award.
The award, formerly called the “Infinite Kilometer Award,” was created in 2012 to highlight the contributions of important members of the MIT science community. The awardees are nominated for their contributions to their research labs, participation in educational programs, exceptional talent, generous NetBet live casinocharacter, service to the community, teamwork, and in general, going above and beyond in their roles at the Institute, especially during the coronavirus pandemic.
The following are the 2021 School of Science Infinite Expansion winners:
- Xinqiang Ding, a postdoc in the Department of Chemistry, nominated by Assistant Professor Bin Zhang for “being one of the most promising, talented, and hard-working scientists that [he has] worked with in [his] entire career”;
- Quentin Ferry, a postdoc in the Picower Institute for Learning and Memory, nominated by Professor Susumu Tonegawa for “remarkable raw talent, versatility … a highly motivated attitude, deep critical thinking, and an extremely creative personality”;
- Hamed Owladeghaffari, a postdoc in the Department of Earth, Atmospheric and Planetary Sciences, nominated by Assistant Professor Matěj Peč for “consistently gone above and beyond his duty”;
- Andrew Grassetti, a postdoc in the Department of Biology, nominated by Assistant Professor Joseph Davis for “[going] well beyond any reasonable expectations to ensure that my entire group has the support — scientific, professional, and emotional — that they needed to succeed”;
- Sarah Heine, a research scientist in the MIT Kavli Institute for Astrophysics and Space Research (MKI), nominated by Principal Research Scientist Herman Marshall for “[being] a major contributor”;
- Samantha Kristufek, a postdoc in the Department of Chemistry, nominated by Professor Jeremiah Johnson for “cultivating an inclusive, supportive group culture”;
- Nathan Lourie, a research scientist in the MIT Kavli Institute for Astrophysics and Space Research, nominated by Professor and MKI Director Rob Simcoe for “demonstrat[ing] both a high degree of personal grit, a capacity to build and lead a team, and a high degree of community engagement”;
- Hiruy Meharena, a postdoc in the Picower Institute for Learning and Memory, nominated by Professor and Picower Institute Director Li-Huei Tsai for “being a community builder and exemplary scientific colleague”;
- Alexander Schuppe, a postdoc in the Department of Chemistry, nominated by Professor Stephen Buchwald for “consistent and significant positive impact on the research efforts of others”;
- Jitendra Sharma, a research scientist in the Picower Institute for Learning and Memory, nominated by administrative manager Eleanor MacPhail, postdoc Grayson Sipe, and Professor Mriganka Sur for “willingness to help everyone,” “serves as a beacon of optimism and collegiality,” and “approach[ing] each day with the goal of making a difference that will help advance the MIT mission”;
- Yong Wang, a postdoc in the Department of Chemistry, nominated by Assistant Professor Alison Wendlandt for “[being] an exceptionally talented scientist, a committed mentor, and a model coworker”;
- Jun Yang, a postdoc in the Department of Physics and MIT Kavli Institute for Astrophysics and Space Research, nominated by Professor Or Hen, professor and physics head Peter Fisher, and Research Scientist Norbert Shulz for “community building,” “mak[ing] a difference,” and “[making] great efforts to organize events for the physics postdoc association during a time of isolation”; and
- Hannah Yevick, a research scientist in the Department of Biology, nominated by Associate Professor Adam Martin for “devotion to mentoring.”
The honor includes a monetary award and will be commemorated in person at a later date with family, friends, and nominators, as well as the winners of the 2021 Infinite Mile Award.

Case’s new lab investigates why cancer arises when disruptions in cellular organization change how cells sense mechanical forces.
Saima Sidik | Department of Biology
February 2, 2021
Assistant professor of biology Lindsay Case wants to understand the protein complexes called focal adhesions that let cells move and sense the world around them. She also aims to determine how cancer arises when focal adhesions malfunction. During her postdoc work at the University of Texas Southwestern Medical Center, she discovered that some of the proteins in focal adhesions work together because of phase separation — a clumping behavior that researchers are just beginning to understand. She sat down to discuss what her work means for cancer research, and her future plans for her new lab in the Department of Biology.
Q: What is phase separation, and how does it affect the way cells function?
A: I always compare phase separation to the separation of oil and vinegar. Something similar can happen with almost any kind of molecule, including proteins. When the interactions between proteins are stronger than their interactions with their surroundings, they can segregate into something called a liquid phase, similar to oil droplets floating in vinegar.
Phase separation matters because organization is a major challenge for our cells, which contain tons of different types of molecules. Sometimes cells organize these molecules using membranes, which is like using fences to keep them in place. But many subcellular structures aren’t surrounded by membranes, and how these compartments keep their components together has been a big mystery since scientists first observed them under microscopes in the 1800s. It’s really only in the last 10 years that people have realized that phase separation is part of the answer.
When cells lose the ability to stay organized, it can have devastating consequences. Changes in how proteins phase separate might underlie serious diseases, like Alzheimer’s and ALS [amyotrophic lateral sclerosis]. I’m really interested in how phase separation organizes protein complexes called focal adhesions, which link cells to their external environment. One function of focal adhesion is to let cells sense mechanical forces from the outside world, and when cells lose this ability it can contribute to cancer progression.
Q: How did phase separation initially pique your interest, and how has your research career prepared you for the work you’ll do at MIT?
A: During my PhD, I was studying how molecules within focal adhesions are organized. I saw a talk by Michael Rosen from the University of Texas Southwestern Medical Center, who would later become my postdoc advisor. Phase separation of proteins was a new idea at the time, but Mike thought it was a powerful force underlying protein organization that we needed to understand more thoroughly. I was intrigued because, at the time, I was unsure what drove focal adhesions to assemble on the plasma membrane, and I wondered if that arrangement might be due to phase separation.
I ended up joining Mike’s lab for my postdoc so that I could apply his ideas about phase separation to my interest in cell signaling and focal adhesions. As a result, I ended up working in a field as it was being born. The first year of my postdoc there were only a few papers investigating phase separation in cellular organization, and now there are over 1,000. Seeing this rapid progress firsthand has been exciting. One of the highlights of my postdoc was showing that phase separation can actually affect the functions of signaling proteins organized on membranes, and I think this discovery went a long way towards showing that phase separation isn’t just a thing that cells can do — it’s something they need to do to survive.
MIT will be an awesome place to continue studying how phase separation lets cells sense the world around them. It’s one of the institutes where the idea of phase separation in biology took off, and the MIT scientists who work on phase separation come from so many different research backgrounds. Understanding phase separation is going to require an interdisciplinary approach, which MIT values. Plus, the students are amazing!
Q: What makes your approach to studying cancer unique?
A: A lot of cancer researchers focus on large-scale or small-scale aspects of these diseases. They either look at how cancer cells behave as a whole, or study the behavior of just one protein. But there’s a level in between where I want to focus my work. I can figure out how large, multi-protein complexes like focal adhesions — some of which form because of phase separation — affect disease progression. During my postdoc, I developed a way to recreate simplified focal adhesions outside of cells. I want to use this system to learn more about how phase separation lets these complexes sense mechanical forces, and how this changes in cancer cells.
Some of the proteins found in focal adhesions are tethered to the plasma membrane, and not many people have studied how protein phase separation changes when you throw a membrane into the mix. I’m excited to keep building up my simplified focal adhesion system in my new lab, and eventually recreate the rest of the complex.
As my lab becomes more established, I’d also like to study how phase separation affects interactions between different protein complexes and signaling pathways. Phase separation is such a rapidly evolving field that it’s hard to know where my research will lead, but that’s part of the fun — not knowing where my work will take me.

“Organs-on-a-chip” system sheds light on how bacteria in the human digestive tract may influence neurological diseases.
Anne Trafton | MIT News Office
January 29, 2021
In many ways, our brain and our digestive tract are deeply connected. Feeling nervous may lead to physical pain in the stomach, while hunger NetBet live casinosignals from the gut make us feel irritable. Recent studies have even suggested that the bacteria living in our gut can influence some neurological diseases.
Modeling these complex interactions in animals such as mice is difficult to do, because their physiology is very different from humans’. To help researchers better understa nd the gut-brain axis, MIT researchers have developed an “organs-on-a-chip” system that replicates interactions between the brain, liver, and colon.
Using that system, the researchers were able to model the influence that microbes living in the gut have on both healthy brain tissue and tissue samples derived from patients with Parkinson’s disease. They found that short-chain fatty acids, which are produced by microbes in the gut and are transported to the brain, can have very different effects on healthy and diseased brain cells.
“While short-chain fatty acids are largely beneficial to human health, we observed that under certain conditions they can further exacerbate certain brain pathologies, such as protein misfolding and neuronal death, related to Parkinson’s disease,” says Martin Trapecar, an MIT postdoc and the lead author of the study.
Linda Griffith, the School of Engineering Professor of Teaching Innovation and a professor of biological engineering and mechanical engineering, and Rudolf Jaenisch, an MIT professor of biology and a member of MIT’s Whitehead Institute for Medical Research, are the senior authors of the paper, which appears today in Science Advances.
The gut-brain connection
For several years, Griffith’s lab has been developing microphysiological systems — small devices that can be used to grow engineered tissue models of different organs, connected by microfluidic channels. In some cases, these models can offer more accurate information on human disease than animal models can, Griffith says.
In a paper published last year, Griffith and Trapecar used a microphysiological system to model interactions between the liver and the colon. In that study, they found that short-chain fatty acids (SCFAs), molecules produced by microbes in the gut, can worsen autoimmune inflammation associated with ulcerative colitis under certain conditions. SCFAs, which include butyrate, propionate, and acetate, can also have beneficial effects on tissues, including increased immune tolerance, and they account for about 10 percent of the energy that we get from food.
In the new study, the MIT team decided to add the brain and circulating immune cells to their multiorgan system. The brain has many interactions with the digestive tract, which can occur via the enteric nervous system or through the circulation of immune cells, nutrients, and hormones between organs.
Several years ago, Sarkis Mazmanian, a professor of microbiology at Caltech, discovered a connection between SCFAs and Parkinson’s disease in mice. He showed that SCFAs, which are produced by bacteria as they consume undigested fiber in the gut, sped up the progression of the disease, while mice raised in a germ-free environment were slower to develop the disease.
Griffith and Trapecar decided to further explore Mazmanian’s findings, using their microphysiological model. To do that, they teamed up with Jaenisch’s lab at the Whitehead Institute. Jaenisch had previously developed a way to transform fibroblast cells from Parkinson’s patients into pluripotent stem cells, which can then be induced to differentiate into different types of brain cells — neurons, astrocytes, and microglia.
More than 80 percent of Parkinson’s cases cannot be linked to a specific gene mutation, but the rest do have a genetic cause. The cells that the MIT researchers used for their Parkinson’s model carry a mutation that causes accumulation of a protein called alpha synuclein, which damages neurons and causes inflammation in brain cells. Jaenisch’s lab has also generated brain cells that have this mutation corrected but are otherwise genetically identical and from the same patient as the diseased cells.
Griffith and Trapecar first studied these two sets of brain cells in microphysiological systems that were not connected to any other tissues, and found that the Parkinson’s cells showed more inflammation than the healthy, corrected cells. The Parkinson’s cells also had impairments in their ability to metabolize lipids and cholesterol.
Opposite effects
The researchers then connected the brain cells to tissue models of the colon and liver, using channels that allow immune cells and nutrients, including SCFAs, to flow between them. They found that for healthy brain cells, being exposed to SCFAs is beneficial, and helps them to mature. However, when brain cells derived from Parkinson’s patients were exposed to SCFAs, the beneficial effects disappeared. Instead, the cells experienced higher levels of protein misfolding and cell death.
These effects were seen even when immune cells were removed from the system, leading the researchers to hypothesize that the effects are mediated by changes to lipid metabolism.
“It seems that short-chain fatty acids can be linked to neurodegenerative diseases by affecting lipid metabolism rather than directly affecting a certain immune cell population,” Trapecar says. “Now the goal for us is to try to understand this.”
The researchers also plan to model other types of neurological diseases that may be influenced by the gut microbiome. The findings offer support for the idea that human tissue models could yield information that animal models cannot, Griffith says. She is now working on a new version of the model that will include micro blood vessels connecting different tissue types, allowing researchers to study how blood flow between tissues influences them.
“We should be really pushing development of these, because it is important to start bringing more human features into our models,” Griffith says. “We have been able to start getting insights into the human condition that are hard to get from mice.”
The research was funded by DARPA, the National Institutes of Health, the National Institute of Biomedical Imaging and Bioengineering, the National Institute of Environmental Health Sciences, the Koch Institute Support (core) Grant from the National Cancer Institute, and the Army Research Office Institute for Collaborative Biotechnologies.

Generous gift from Michael Gould and Sara Moss provides endowed support for MIT’s Summer Research Program in Biology.
Department of Biology
January 27, 2021
Last month, the Department of Biology received a generous gift from Michael Gould and Sara Moss to support students participating in MIT’s Summer Research Program in Biology (MSRP-Bio). Gould is a philanthropist and the retired chair and CEO of Bloomingdales, and Moss is the vice chair of Estée Lauder Companies. Their gift will supplement the existing Bernard S. and Sophie G. Gould Fund, which the couple initiated in 2015 to honor Gould’s parents. Together, these donations will enable many undergraduate students from outside MIT who are interested in a career in life science to participate in MSRP-Bio each summer. To honor the Gould family’s generosity, MSRP-Bio will be renamed the Bernard S. and Sophie G. Gould MIT Summer Research Program in Biology, or BSG-MSRP-Bio.
“We are deeply grateful to Mike and Sara for their commitment to and support for our community,” says department head and Praecis Professor Alan Grossman. “Their willingness to enable opportunities for students will allow many talented individuals to benefit from research experiences here at MIT, and foster the next generation of scientists.”
“Mike Gould and Sara Moss are amazing people,” says Mandana Sassanfar, the Department of Biology’s director of outreach. “They’ve made a generous gift that has enabled MRSP-Bio to give many deserving undergraduates a life-changing summer research experience.”
MSRP-Bio is a 10-week summer program offered to non-MIT undergraduates, which provides access to cutting-edge facilities and supervised research in a fast-paced environment. The program encourages students from groups that are historically underrepresented in science, first-generation college students, students from economically disadvantaged backgrounds, and students with disabilities to attend graduate school and pursue careers in basic research.
Every year, roughly 20 participants are placed in laboratories affiliated with the Department of Biology. In total, nearly 400 students have participated in MSRP-Bio since its establishment in 2003. Nearly 300 have already gone to PhD or MD/PhD programs, and of these, 63 have enrolled at MIT for graduate school and 45 have joined the Department of Biology specifically.
Gould and Moss were inspired to create a fund supporting MSRP-Bio because both of Gould’s parents were MIT alumni devoted to mentorship. Bernard “Bernie” Gould ’32 was a beloved biochemistry professor in the Department of Biology and counseled many biology and pre-med students for 50 years. His wife, Sophia Gould CMP ’48, earned a master’s degree in public health at a time when there were few female netbet online sports bettinggraduate students at the Institute, and shared this passion for training students.
“I’ve been inspired by my father, who was a first-generation American, cared enormously about his students, and nurtured their intellectual curiosity and drive,” Michael Gould says. “MSRP-Bio does the same by giving each student the opportunity of a lifetime. My dream is for every department at MIT to create a similar program. It would enrich the Institute immeasurably.”
“This gift will have a tremendous impact on the MSRP-Bio program in the biology department, and comes at a crucial time as issues surrounding diversity, equity, and inclusion remain key priorities for the School of Science and Institute,” says Nergis Mavalvala, dean of MIT’s School of Science.
Since its inauguration in 2015, the Bernard S. and Sophie G. Gould Fund has offered scholarships to 30 MSRP-Bio students — six each year. Every summer, Gould and Moss travel to campus to get to know the current Gould fellows. The duo has been continually impressed by the caliber of students, and decided to provide more support to fund additional fellows and ensure the program’s longevity.
“Mike and Sara have a sustained interest in the well-being and success of the Gould fellows, and take pride in these students’ accomplishments,” Sassanfar says. “The Gould Fellowship stands out because of the open relationship between the Mike and Sara and the fellows, which forges meaningful connections that will last for many years.”
Participating in MSRP-Bio was a “life-changing” experience, says former Gould Fellow Meucci Ilunga. “I genuinely mean that — I can only imagine how different my life would be if I had not had that opportunity.”
Former Gould Fellow Asmita Panthi adds that MSRP-Bio showed her what she was capable of accomplishing, and gave her the confidence to apply to graduate school. “I’m so thankful for this impactful fellowship, which gives students like me — who come from small undergraduate institutions or humble backgrounds — the chance to participate in a rigorous research program.”

Neurons dynamically control their myelin patterns.
Picower Institute
January 1, 2021
Harvard and MIT researchers have discovered a new way that the brain responds to stimuli, with different types of neurons using myelin in different ways. By dynamically controlling myelin, the insulating coating around their long axon projections that helps with signal conduction, neurons have more ways to adapt to changes. Published in the journal Science, the study in mice advances scientists’ understanding of how the brain works and opens avenues for exploring new disease mechanisms.
“The brain requires a diversity of mechanisms at its disposal in order to adapt to stimuli. We know that neurons have different properties, and we demonstrated several years ago that neurons have different patterns of myelination. Now, we have found an additional layer of complexity: neurons actively use their myelin in dynamic and different ways,” said co-senior author Paola Arlotta, the Golub Family Professor of Stem Cell and Regenerative Biology at Harvard University.
In biology textbooks, the prototypical image of a neuron depicts the axon with a series of equally sized, evenly spaced pieces of myelin. However, the Arlotta lab showed in 2014 that the picture is more complicated: different types of neurons show different patterns of myelination, with varying lengths of myelin or no myelination on some segments. In the current study, the researchers delved deeper into the phenomenon of how myelination patterns might change over time.
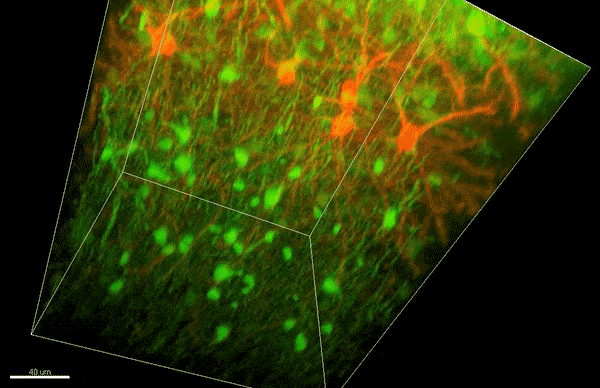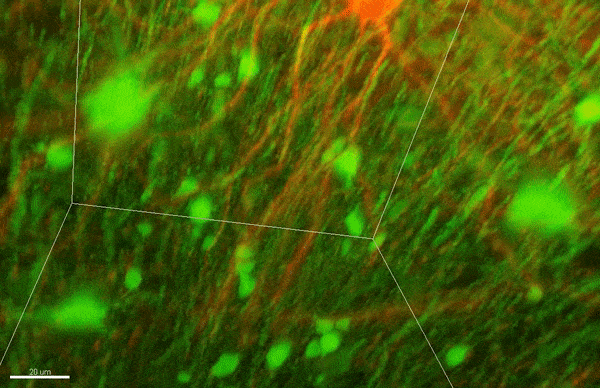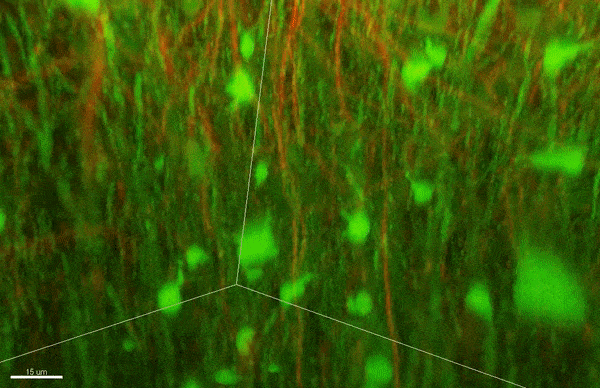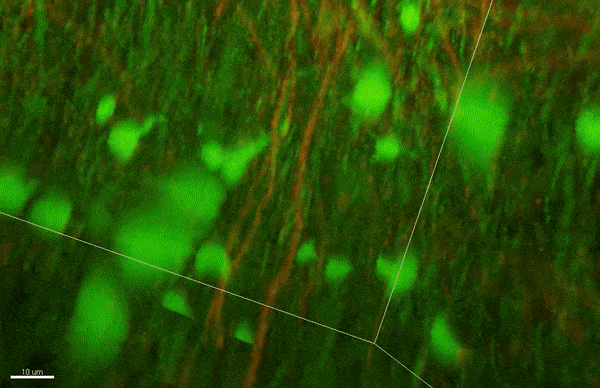
To investigate myelin plasticity, the researchers used mouse models where specific neuron types were fluorescently labeled. They changed the animals’ sensory input by closing one eye, then tracked how the brain responded using a custom-built in vivo imaging system in the lab of collaborator and co-senior author Elly Nedivi.
“Our multicolor method enables the simultaneous visualization of both the myelin and the axons it was wrapping. This allowed us to closely track how myelin was changing over time as mice reconfigured the visual cortex as sight became deprived in one eye,” said Nedivi, who is the William R. (1964) & Linda R. Young Professor of Neuroscience at MIT, and a member of The Picower Institute for Learning and Memory and the Departments of Biology and Brain and Cognitive Sciences.
The researchers found that even though they tracked neurons that were next to each other and part of the same network, different cell types had different responses — specifically, inhibitory neurons remodeled their myelin more than excitatory neurons.
“In the inhibitory neurons, we saw a two-times increase in the number of myelin changes. Those changes can be any way you can imagine: they can be myelin segments shortening or elongating, the addition of new myelin, and also the elimination of an entire piece of myelin,” said Sung Min Yang, lead author and postdoctoral fellow in the Arlotta lab.
The unique capacity to change their myelin opens up possibilities for the neurons, Yang said: “It turns out that neurons do not move myelin around in a consistent way, as in a game of checkers where every game piece has the same move. Instead, the brain is playing chess, where different neurons — or pieces — can move in different ways. This gives the brain more choice in how to use myelin, which is a limited resource.”
The researchers also found that neurons did not necessarily have to produce new myelin, but could reuse and reshape what they already had in order to respond.
“We have discovered a fundamental property of the brain that opens the window to conceptualizing how the organ maximizes its power and optimizes its function. The dynamic distribution of myelin is yet another level of mechanism that the brain uses to diversify its response to a given stimulus — the endless combinations can enable a more complex, even surprising outcome,” Arlotta said.
Based on the findings, the researchers can now investigate how myelin plasticity plays a role in other contexts, including disease.
“We hope to be able to investigate myelin pathology in human brain organoid models, which can be generated from patients or engineered to contain specific mutations associated with myelin abnormalities, in order to better understand the disease mechanisms,” Arlotta said.
This research was supported by the Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard, the National Institute of Mental Health, the National Institutes of Health, and the JPB Foundation.

Pablo Jarillo-Herrero, Aviv Regev, Susan Solomon, and Feng Zhang are the recipients of distinguished awards for major contributions to science.
Laura Carter | School of Science
January 25, 2021
Four MIT scientists are among the 20 recipients of the 2021 Academy Honors for major contributions to science, the National Academy of Sciences (NAS) announced at its annual meeting. The individuals are recognized for their “extraordinary scientific achievements in a wide range of fields spanning the physical, biological, social, and medical sciences.”
The awards recognize: Pablo Jarillo-Herrero, for contributions to the fields of nanoscience and nanotechnology through his discovery of correlated insulator behavior and unconventional superconductivity in magic-angle graphene superlattices; Aviv Regev, for using interdisciplinary information or techniques to solve a contemporary challenge; Susan Solomon, for contributions to understanding and communicating the causes of ozone depletion and climate change; and Feng Zhang, for pioneering achievements developing CRISPR tools with the potential to diagnose and treat disease.
Pablo Jarillo-Herrero: Award for Scientific Discovery
Pablo Jarillo-Herrero, a Cecil and Ida Green Professor of Physics, is the recipient of the NAS Award for Scientific Discovery for his pioneering developments in nanoscience and nanotechnology, which is presented to scientists in the fields of astronomy, materials science, or physics. His findings expand nanoscience by demonstrating for the first time that orientation can be used to dramatically control nanomaterial properties and to design new nanomaterials. This work lays the groundwork for developing a whole new family of 2D materials and has had a transformative impact on the field and on condensed-matter physics.
The biannual award recognizes “an accomplishment or discovery in basic research, achieved within the previous five years, that is expected to have a significant impact on one or more of the following fields: astronomy, biochemistry, biophysics, chemistry, materials science, or physics.”
In 2018, his research group discovered that by rotating two layers of graphene relative to each other by a magic angle, the bilayer material can be turned from a metal into an electrical insulator or even a superconductor. This discovery has fostered new theoretical and experimental research, inspiring the interest of technologists in nanoelectronics. The result is a new field in condensed-matter physics netbet sports bettingthat has the potential to result in materials that conduct electricity without resistance at room temperature.
Aviv Regev: James Prize in Science and Technology Integration
Aviv Regev, who is a professor of biology, a core member of the Broad Institute of Harvard and MIT, a member of the Koch Institute, and a Howard Hughes Medical Institute investigator has been selected for the inaugural James Prize in Science and Technology Integration, along with Harvard Medical School Professor Allon Kelin, for “their concurrent development of now widely adopted massively parallel single-cell genomics to interrogate the gene expression profiles that define, at the level of individual cells, the distinct cell types in metazoan tissues, their developmental trajectories, and disease states, which integrated tools from molecular biology, engineering, statistics, and computer science.”
The prize recognizes individuals “who are able to adopt or adapt information or techniques from outside their fields” to “solve a major contemporary challenge not addressable from a single disciplinary perspective.”
Regev is credited with forging new ways to unite the disciplines of biology, computational science, and engineering as a pioneer in the field of single-cell biology, including developing some of its core experimental and analysis tools, and their application to discover cell types, states, programs, environmental responses, development, tissue locations, and regulatory circuits, and deploying these to assemble cellular atlases of the human body that illuminate mechanisms of disease with remarkable fidelity.
Susan Solomon: Award for Chemistry in Service to Society
Susan Solomon, the Lee and Geraldine Martin Professor of Environmental Studies in the Department of Earth, Atmospheric and Planetary Sciences who holds a secondary appointment in the Department of Chemistry, is the recipient of the Award for Chemistry in Service to Society for “influential and incisive application of atmospheric chemistry to understand our most critical environmental issues — ozone layer depletion and climate change — and for her effective communication of environmental science to leaders to facilitate policy changes.”
The award is given biannually for “contributions to chemistry, either in fundamental science or its application, that clearly satisfy a societal need.”
Solomon is globally recognized as a leader in atmospheric science, notably for her insights in explaining the cause of the Antarctic ozone “hole.” She and her colleagues have made important contributions to understanding chemistry-climate coupling, including pioneering research on the irreversibility of global warming linked to anthropogenic carbon dioxide emissions, and on the influence of the ozone hole on the climate of the southern hemisphere.
Her work has had an enormous effect on policy and society, including the transition away from ozone-depleting substances and to environmentally benign chemicals. The work set the stage for the Paris Agreement on climate, and she continues to educate policymakers, the public, and the next generation of scientists.
Feng Zhang: Richard Lounsbery Award
Feng Zhang, who is the James and Patricia Poitras Professor of Neuroscience at MIT, an investigator at the McGovern Institute for Brain Research and the Howard Hughes Medical Institute, a professor of brain and cognitive sciences and biological engineering at MIT, and a core member of the Broad Institute of MIT and Harvard, is the recipient of the Richard Lounsbery Award for pioneering CRISPR-mediated genome editing.
The award recognizes “extraordinary scientific achievement in biology and medicine” as well as stimulating research and encouraging reciprocal scientific exchanges between the United States and France.
Zhang continues to lead the field through the discovery of novel CRISPR systems and their development as molecular tools with the potential to diagnose and treat disease, such as disorders affecting the nervous system. His contributions in genome engineering, as well as his earlier work developing optogenetics, are enabling a deeper understanding of behavioral neural circuits and advances in gene therapy for treating disease.
In addition, Zhang has championed the open sharing of the technologies he has developed through extensive resource sharing. The tools from his lab are being used by thousands of scientists around the world to accelerate research in nearly every field of the life sciences. Even as biomedical researchers around the world adopt Zhang’s discoveries and his tools enter the clinic to treat genetic diseases, he continues to innovate and develop new technologies to advance science.
The National Academy of Sciences is a private, nonprofit society of distinguished scholars, established in 1863 by the U.S. Congress. The NAS is charged with providing independent, objective advice to the nation on matters related to science and technology as well as encouraging education and research, recognize outstanding contributions to knowledge, and increasing public understanding in matters of science, engineering, and medicine. Winners received their awards, which include a monetary prize, during a virtual ceremony at the 158th NAS Annual Meeting.
This story is a modified compilation from several National Academy of Sciences press releases.

Eva Frederick | Whitehead Institute
January 21, 2021
Sex-specific chromosomes are a dangerous place to be, if you’re a gene. Because these chromosomes — Y chromosomes, in humans — do not have a matching chromosome with which to exchange genetic information, they are prone to losing non-essential genes left and right in a process called genetic decay.
Now, a new study from research scientist Daniel Winston Bellott in the lab of Whitehead Institute Member David Page broadens our understanding of what makes a gene able to survive on a sex-specific chromosome by looking at one especially slithery branch of the evolutionary tree: snakes.
Comparing surviving genes on snake sex-specific chromosomes to those that are lost to the ravages of time can teach scientists about the evolutionary pressures that shaped sex chromosomes as we know them today. “You might think, ‘These are sex chromosomes, so the surviving genes should have something to do with sex, right?’” Bellott said. “But they don’t.”
Instead, many of these genes are essential to the survival of the animal, and take part in key developmental processes. “It turns out that these survivor genes on sex-specific chromosomes may play a very big role in governing how all of the genes across all the chromosomes are read, interpreted and expressed,” said Page, who is also a professor of biology at Massachusetts Institute of Technology (MIT) and an Investigator of the Howard Hughes Medical Institute. “Winston’s study is absolutely foundational to our understanding of what the sex chromosomes are, how the two sexes come to be, and how health and disease traits play out similarly or differently in males and females.”
What is a sex chromosome, anyway?
Over the course of evolution, all sex chromosomes start out as regular, matching chromosomes called autosomes. Then, somewhere along the line, a mutation happens, and one of the chromosomes gains a “switch,” that, when present, causes an embryo to to develop as a specific sex. “It’s actually really easy to make a sex chromosome,” said Bellott. “In most cases, you only need to change one or two genes and you’ve started the sex chromosome system.”
This process has happened numerous times during the course of evolution. It makes sense; sexual reproduction is an efficient way to ensure genetic diversity. But the whole thing is a bit mysterious; are, for example, certain chromosomes predisposed to become sex chromosomes?
That’s where Bellott thought snakes could be especially helpful. “Snakes have a relatively old system of sex chromosomes, where you have a lot of time for the chromosomes to diverge,” Bellott said. “Time has swept away all the genes that aren’t important, and you can see what kind of genes are left.”
Their sex chromosome system also evolved from different autosomes, some 100 million years after humans’, and thus would provide a useful vantage point from which to consider our own genomes.
To learn more about the evolution of these chromosomes, Bellott and Page first gathered a list of “ancestral genes,” which were likely on the chromosome from which the snake sex chromosomes evolved. New sequencing data for several species of animals distantly related to snakes meant that they had a more complete list of these genes — 1,648 to be exact.
Bellott began painstakingly sifting through the genes that remained on the sex-specific chromosomes of three species of snake: the pygmy rattlesnake, mountain garter snake, and the five-pacer viper. He eventually identified 103 ancestral genes that had survived as long as 90 million years of evolution on the snakes’ sex chromosomes. With this list in hand, Bellott could then ask what these surviving genes had in common that set them apart from the hundreds of genes that were swept off the snakes’ sex chromosomes by genetic decay.
What makes a survivor?
To Bellott’s surprise, the genes that remained on the snakes’ sex-specific chromosome had nothing to do with sex determination; neither were they expressed more often in sex-specific tissues, or more often in one sex than the other.
Instead, Bellott and Page’s research identified three key properties that led to a gene’s survival on snake sex-specific chromosomes. First, the gene must be dosage sensitive. In other words, the snake’s body depends on its cells to produce an exact amount of that gene’s protein product. Any more, or any less, and the snake experiences illness or death. Second, a surviving gene is likely broadly expressed in different tissues across the body, not localized to one specific organ or area. And third, surviving genes are subject to strong purifying, or negative, selection. Simply put, this means that if something goes wrong with one of these genes, the snake has a slim chance of survival or producing offspring.
When Bellott dove deeper into the genes’ function, he discovered that for many of them the equivalent gene in humans played a role in key developmental processes such as the formation of the face. When these genes were mutated in humans, their faces — and other essential parts of the body — would not develop properly. “What Winston is seeing here is that the genes that were preserved on the sex specific chromosomes in snakes are disproportionately involved in birth defects in people,” Page said. “We think that nature is selecting for the survival of [sex chromosome] genes whose dosage in certain parts of embryonic development is especially critical.”
In time, Bellott said, this may allow scientists to predict genes whose role in developmental disorders is yet to be discovered. “In some sense, you get to the place where you’re starting to work the experiment backwards in your mind, and say, ‘Let’s take the set of genes that are on sex specific chromosomes in snakes and birds, but that have not yet been implicated in birth defects in humans,’” Page said. “They might be prime candidates to be responsible for heretofore unexplained birth defects.”
From snakes to humans
Next, the researchers sought to broaden their scope. They compared ancestral genes across the three species of snakes and 38 species of birds and mammals with a larger pool of genes that made it to the present day. Many of the surviving genes on bird and mammal chromosomes had different functions than those on snake chromosomes, but again, most had little to do with sex determination.
“Adding the snakes in with the birds and the mammals gave Winston enough data points to be able to see further and to see more precisely, and now for the first time, he was able to confirm something that we had been suspecting for a long time but really didn’t have sufficient data to pin down,” Page said. “And that is that the chromosomes that became sex chromosomes were not sort of inclined to function in sex differences. Before they got picked out of the crowd, they weren’t specialized towards differentiating between the sexes in much of any way.”
Then, as genes were lost over time, evolutionary pressures ensured that the same sort of genes survived. This idea that sex chromosomes — besides their key developmental switch — have little do do with sex determination challenges the common notion of what a sex chromosome actually is.
“I hope people will pick up on this idea that the chromosomes that became sex chromosomes weren’t in any way preordained,” Page said. “They were just ordinary chromosomes out for a walk in the park, and something happened.”
In the future, Bellott and Page plan to further broaden their scope to include other animals, toward the ultimate goal of understanding our own sex chromosomes. “We take these results, and we turn them into a lens through which we look at sex differences in health and disease in our own species,” Page said. “This research really refines our ideas about what it means to be a gene on the human X or Y chromosome, and how we should think about those genes that survive.”
Bellott, D.W. and Page, D.C. “Dosage-sensitive functions in embryonic development drove the survival of genes on sex-specific chromosomes in snakes, birds, and mammals.” Genome Research, Jan. 21, 2021. DOI: 10.1101/gr.268516.120.

Eva Frederick | Whitehead Institute
January 21, 2021
When cancer is confined to one spot in the body, doctors can often treat it with surgery or other therapies. Much of the mortality associated with cancer, however, is due to its tendency to metastasize, sending out seeds of itself that may take root throughout the body. The exact moment of metastasis is fleeting, lost in the millions of divisions that take place in a tumor. “These events are typically impossible to monitor in real time,” said Whitehead Institute Member Jonathan Weissman.
Now, researchers led by Weissman, who is also a professor of biology at Massachusetts Institute of Technology and an investigator with the Howard Hughes Medical Institute, have turned a CRISPR tool into a way to do just that. In a paper published January 21 in Science, Weissman’s lab, in collaboration with Nir Yosef, a computer scientist at the University of California, Berkeley, and Trever Bivona, a cancer biologist at the University of California, San Francisco (UCSF), treats cancer cells the way evolutionary biologists might look at species, mapping out an intricately detailed family tree. By examining the branches, they can track the cell’s lineage to find when a single tumor cell went rogue, spreading its progeny to the rest of the body.
“With this method, you can ask questions like, ‘How frequently is this tumor metastasizing? Where did the metastases come from? Where do they go?’” Weissman said. “By being able to follow the history of the tumor in vivo, you reveal differences in the biology of the tumor that were otherwise invisible.”
Scratch paper cells
Scientists have tracked the lineages of cancer cells in the past by comparing shared mutations and other variations in their DNA blueprints. These methods, however, depend to a certain extent on there being enough naturally occurring mutations or other markers to accurately show relationships between cells. That’s where Weissman and co-first authors Jeffrey Quinn, then a postdoctoral researcher in Weissman’s lab, and Matthew Jones, a graduate student in Weissman’s lab, saw an opportunity to use CRISPR technology — specifically, a method developed by Weissman Lab member Michelle Chan to track embryo development — to facilitate tracking. Instead of simply hoping that a cancer lineage contained enough lineage-specific markers to track, the researchers decided to use Chan’s method to add in markers themselves. “Basically, the idea is to engineer a cell that has a genomic scratchpad of DNA, that then can be ‘written’ on using CRISPR,” Weissman said. This ‘writing’ in the genome is done in such a way that it becomes heritable, meaning a cell’s grand-offspring would have the ‘writing’ of its parent cells and grandparent cells recorded in its genome.
Notes
Jeffrey J. Quinn, Matthew G. Jones, Ross A. Okimoto, Shigeki Nanjo, Michelle M. Chan, Nir Yosef, Trever G. Bivona, Jonathan S. Weissman. “Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts.” Science, Jan. 21, 2021.
